Next-Generation Sequencing (NGS) Bioinformatic Pipelines
Bioinformatic pipeline and data processing descriptions, workflow steps, quality control analysis,
and available client data and reports. Champions data pipelines are run and monitored in the
Amazon Elastic Compute Cloud environment.
Current Pipelines Available: RNAseq, Whole-Exome Sequencing (WES)
Data Pipelines Workflow Overview:
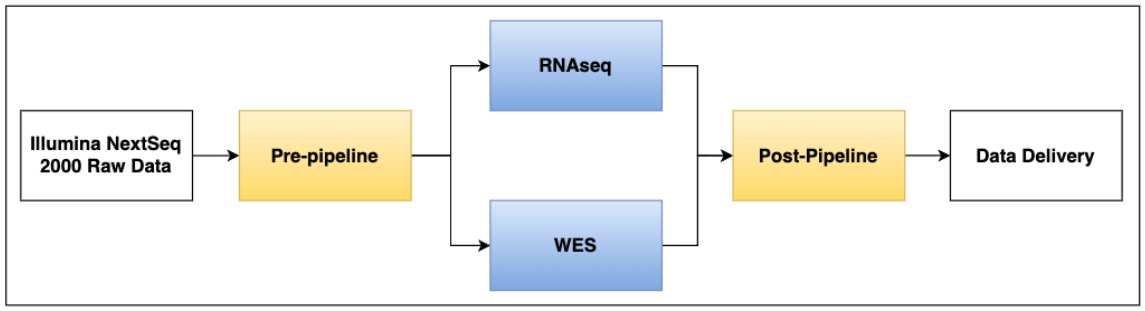
Workflow Overview
Illumina-Demultiplex Pre-Pipeline
The illumina-demultiplex Nextflow pipeline serves as the ‘Pre-pipeline’ after the wet-lab and inhouse
(RNAseq or WES) sequencing is complete. Human or mouse sequencing raw data and a valid
sample sheet can be used as input.
Steps:
- Data from the sequencing instrument is transferred to the AWS cloud environment.
- The raw data from the Illumina NextSeq 2000 instrument (*.bcl files) is used to convert to
generate fastq.gz files. - The fastq.gz files undergo an extensive quality control process in which the sequence PHRED
quality score, %PF (reads passing filter), read indexing score, and read count threshold, must
meet a minimum threshold to pass QC. - Fastq.gz sequence files that pass QC will then undergo a concatenation (if multiple duplicate
sample IDs exist) step if necessary. - A pipeline run manifest for RNAseq, or WES, is generated and used to launch each respective
pipeline.
Reports/Output Available for Clients:
- Fastq.gz Quality Control Report.
RNAseq Pipeline
The RNAseq Nextflow pipeline uses known bioinformatic tools to analyze and aggregate data from
fastq.gz sequence files. This pipeline accepts PDX (which will be de-moused in silico) or human
samples. The raw data generated in the RNAseq pipeline undergoes post-pipeline processing to
output normalised Expression of gene transcripts, and gene Fusions.
Steps:
- QC of fastq files using FastQC.
- Fusion analysis using FusionCatcher with the most up to date human genome references.
- Trimming: Fastq files are trimmed for low quality sequences and sequencing adaptors.
- Alignment: After trimming, fastq sequence files are aligned to a combined index of human
and mouse reference genomes (GRCh37 and GRCm38) using STAR aligner (v2.4.2a). - De-mousing: Fastq files undergo a de-mousing step, in which the mouse ‘host’ is removed.
- Alignment: Alignment of fastq to bacterial and viral genomes. This step is to check if both
bacteria and/or viruses are present. - Expression quantification using FeatureCount (v1.4.3) and RSEM.
- Sample Haplotype: Derive a sample haplotype against a given VCF file.
- QC of data generated using Picard (v1.83), as well as de-mousing statistics.
Reports Available for Clients:
- RNAseq QC Metrics Summary Report
Whole-Exome Sequencing (WES) Pipeline
The Whole-exome sequencing (WES) Nextflow pipeline uses the well-known GATK pipeline for local
realignment around INDELs and base quality score recalibration, as well as SNP analysis using
multiple callers (muTect, Lofreq and Strelka) against a normal cell line sample as a control
(NA12878). This pipeline accepts PDX (which will be de-moused) or human samples. The raw data
generated in the WES pipeline is processed to produce filtered Mutations (SNVs and INDELs), Copy
Number Variants (CNV), MSI and TMB (tumor mutation burden) score and HLA typing.
Steps:
- Trimming: Fastq files are trimmed for low quality sequences and sequencing adaptors.
- AGeNT trimming and FastQC: For SureSelect XT HS2 libraries, in which a molecular tag is
added to monitor read duplication rate, then followed up by FastQC of the tagged Fastq files. - Alignment: Alignment is performed using BWA aligner using a combined index of human and
mouse reference genomes (GRCh37 and GRCm38). - De-mousing: combined BAM files undergo a de-mousing step, in which the mouse ‘host’ is
removed. - Alignment: Alignment is performed again using BWA aligner.
- Mark duplicates. For samples generated with Agilent library prep kit and human only
samples. - De-duplication: De-duplication to remove duplicate reads.
- INDEL realignment: GATK3 is used for INDEL realignment.
- Add Read Groups: Read groups are added after INDEL realignment using Picard Tools.
- Base recalibration: GATK3 base recalibration is performed on post-alignment output (bam).
- Clip overlap: Overlapping reads are clipped in read pairs of post-alignment output (bam) of
both normal samples AND the tumor ‘normal’ sample included on the run. - Collect exon metrics: Exon metrics are collected using Picard ‘CollectHsMetrics’ function and
using the latest human reference sequence. - Call INDELS: Call INDELs using Scalpel v0.5.4., Strelka v2.9.10., and Pindel v0.2.5b9.
- Call SNVs: Call SNV using Lofreq v2.1.4., MuTect v1.1.7., Strelka v2.9.10., and Pindel v0.2.5b9.
- Post-processing: GATK3 for variant selection and filtration.
- Combine Variants: Combine filtered VCF (variant calling file) files from different callers.
- Annotate combined VCF file using SnpSift and VEP
- Detect microsatellite instability: MSISensor2 for microsatellite instability detection.
- CNV: Detect copy number variations with EXCAVATOR2 (v1.1.2).
- HLA Typing: HLA typing performed using Optitype.
- Sample Haplotype: Derive a sample haplotype against a given VCF file.
Reports Available for Clients:
- WES QC Metrics Summary Report
Post-Pipeline QC & Data Processing
Quality Control and data processing is run after each run of RNAseq or WES. All data generated in
each respective pipeline undergoes extensive QC and data processing prior to releasing to a client
or uploading to our Lumin Bioinformatics platform. Each instance of post-pipeline data-processing is
unique for each pipeline and produces unique output types.
Steps:
- Aggregation of output data from pipeline run.
- QC of aggregated data.
- Data that passes QC will undergo further data processing using in-house scripts to produce
the following final data output:
RNAseq:
-
- Expression
-
- Fusion
-
- Haplotype
WES:
-
- Mutations
-
- Copy Number Variations (CNV)
-
- HLA Typing
-
- MSI Scores
-
- TMB (tumor mutation burden)
-
- Haplotypes
- Final processed data is prepared for delivery to the client or is prepared for upload to our
Lumin Bioinformatics platform.
Updated almost 3 years ago
