PDX
Champions derived PDX models
The Champions PDX bank was derived from human patient tumors prior to (treatment-naïve) or after undergoing standard-of-care therapy (post-therapy). It is a collection of 1000+ low-passage PDXs (patient-derived xenograft) derived from human tumor samples.
As part of our internal research and development, we performed over 3,400 in-vivo PDX efficacy studies and obtained response rates of our PDX models to a number standard of care therapies. Molecular data (whole-exome and RNA sequencing) from these PDXs was obtained after passaging (2-3 passages) in mice. This gene signature was calculated based on the differential expression of the user specified gene in PDXs obtained from post-therapy vs. treatment-naïve patient tumors. Thus, this signature reflects the gene expression landscape of the indicated tumors after undergoing standard-of-care therapy with the indicated drugs. The individual values are sign-corrected log10 p-values of gene expression differences. E.g. a value of -2, (representing a p-value of 0.01), for gene “A” indicates that the expression of gene “A” is reduced in patient cancer samples after receiving the standard-of-care therapies described relative to treatment-naïve samples. Conversely, a value of 2, (representing a p-value of 0.01), indicates increased expression of gene “A” in patient cancer samples after receiving the standard-of-care therapies described relative to treatment-naïve samples.
Many of these are the same models available to clients to commission their own studies.
PDX update!
In addition to PDX RNASeq, Champions now offers PDX proteomics, PDX phosphoproteomics, and PDX Kinase for the Clustergrammer and Networks visualizations.
Model Select Page
View the status of sequencing and proteomics in ever updating bank of PDX models on our Model Select Page. The guide for using this can be found under 'Lumin Tools'.
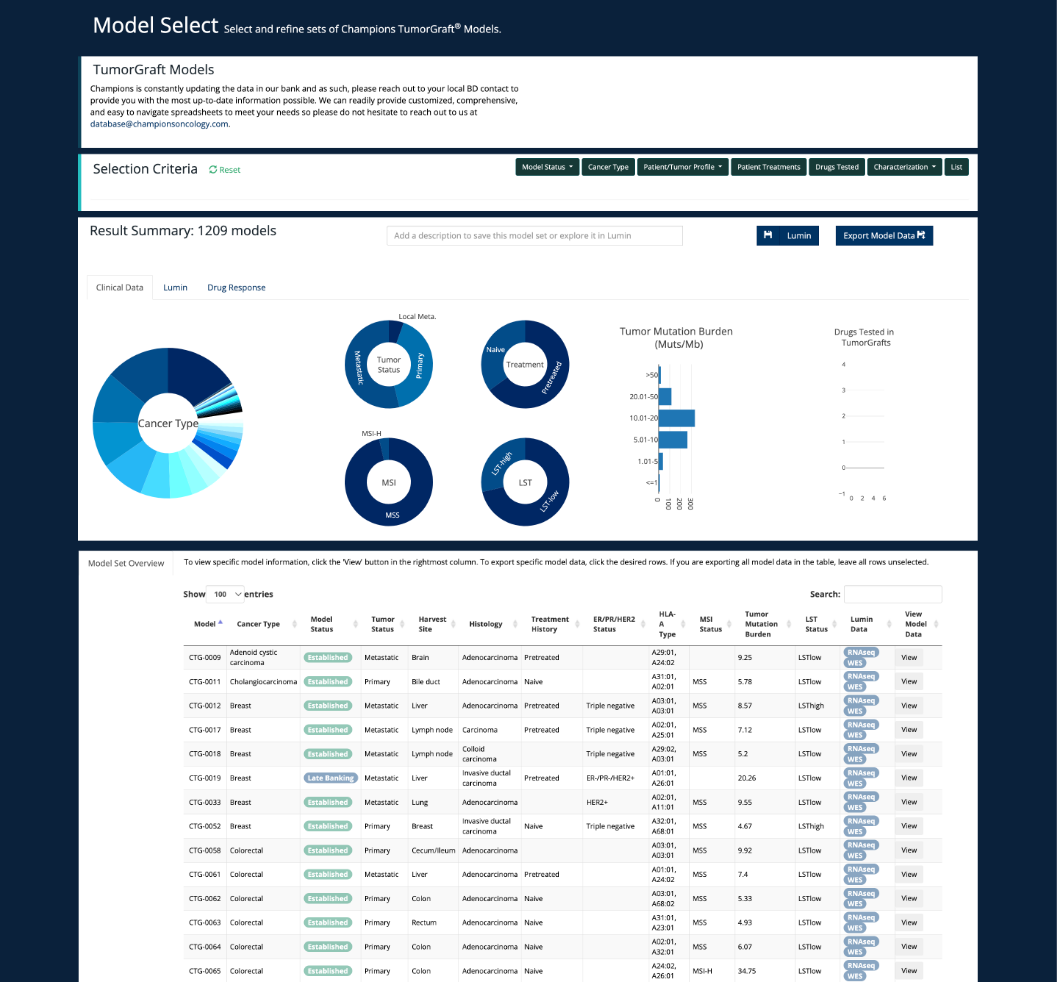
Please find more information on this under 'Lumin Tools - Model Select'.
Updated over 2 years ago
