Clustering Heatmap
Explore large datasets with Champions' Clustering Heatmap.
General Description
Our Clustering Heatmap is a web-based tool which aims to visualize and analyze high-dimensional data as interactive and shareable hierarchically clustered heatmaps. Build heatmaps using Champions multiomic PDX dataset or customize gene expression heatmaps using public datasets, Champions cell lines or your own in-house datasets.
Clustergrammer Inputs
Clustergrammer Inputs:
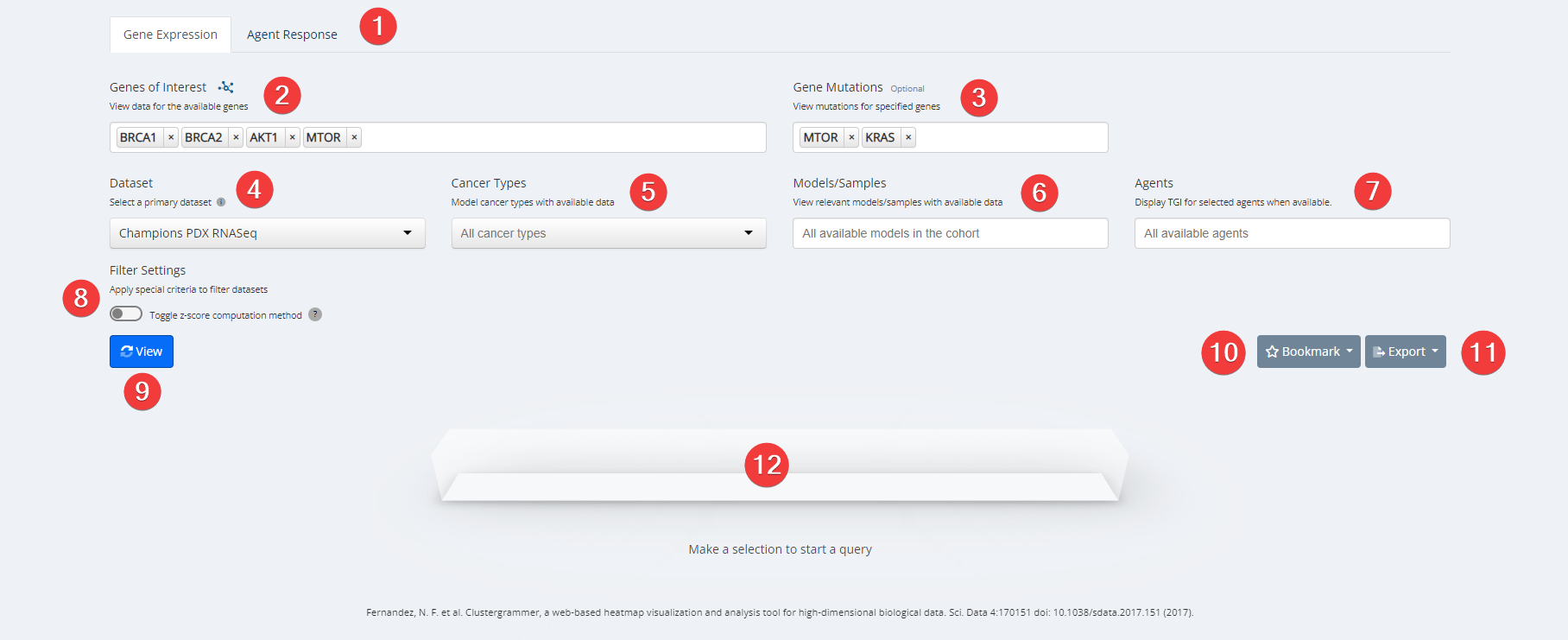
Clustergrammer Inputs
- Analysis Type. Choose between Gene Expression and Agent Response.
- Genes of Interest. Select your genes or proteins of interest. Only options with available data for your other selections will be clickable. Options that are greyed out have no available data for your other selections.
- Gene Mutation. View mutations for specified genes. Only options with available data for your other selections will be clickable. Options that are greyed out have no available data for your other selections.
- Dataset. Choose from the dropdown menu between Champions Database, Online Database such as TCGA, GEO, CPTAC, or your own data previously uploaded to Lumin.
- Cancer Type. Select cancer type of interest from the dropdown menu or leave blank for all cancer types. Please Note: A specific cancer type is required for all datasets other than Champions PDX and TCGA.
- Model/Sample. Select specific models with available data or leave blank for all available models with data.
- Agents. Select specific test agents with available data or leave blank for all available test agents with data.
- Filter Settings.
- a) Champions PDX RNASeq and TCGA Datasets - Toggle between calculating z-scores for the selected criteria (left) or using precomputed scores across the entire available dataset (right). By default z-scores are computed for the selected criteria.
- b) Champions PDX Proteomics and Phosphoproteomics Datasets - Toggle between using lenient (left) or raw unfiltered intensity scores (right). By default lenient scores are used.
- View.
- Bookmark. Save an analysis or load from an existing analysis. Please Note: As we are continually updating our data the visualization may change slightly from the time it was first analyzed. Users can also manage all bookmarks under "My Bookmarks" in Account Settings at the top right of any Lumin page.
- Export. Analysis can be exported as a CSV file, PNG image or into Lumin Workspaces.
- Loading Task Bar. Building your requested visualization may take some time. The status of your request will be displayed here.
Clustergrammer Visualization
Heatmap for genes BRCA1, BRCA2, AKT1, MTOR with gene mutations for MTOR and KRAS in the Champions PDX dataset with all cancer types, models and agents visualized.
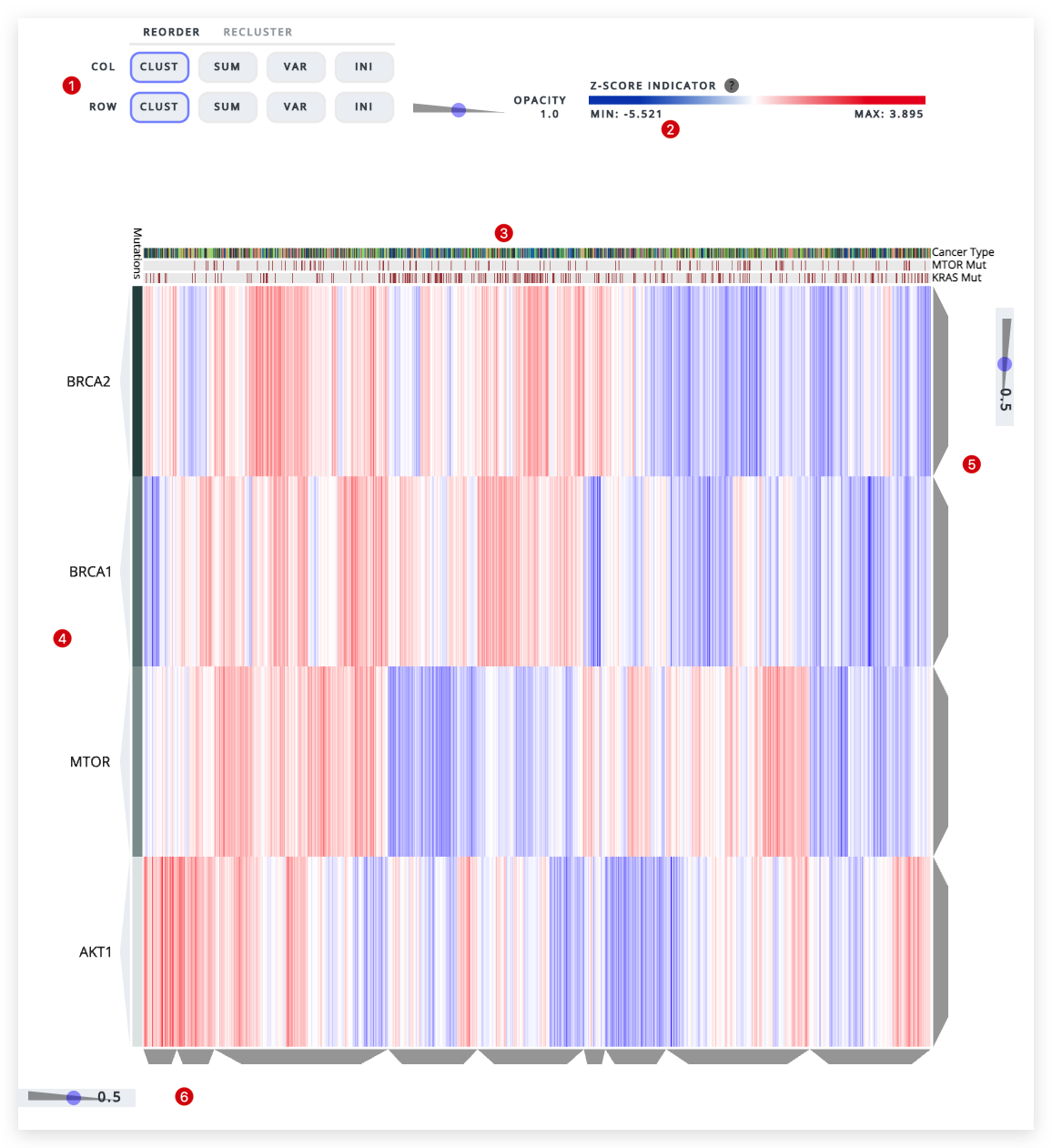
Clustergrammer Visualization
- Heatmap Sorting. Sort/Reorder rows and columns based on the options within each tab.
- Legend. Shows the minimum (blue) to maximum (red) z-score values.
- Y-Axis. Lists genes or proteins that were entered into the 'Genes of Interest' input.
- X-Axis. Displays cancer types and models selected, as well as any gene mutations which have been inputted.
- Dendrogram-Slider Y-Axis. Use to adjust dendrogram cluster sizes - slide up for larger clusters and down for smaller clusters.
- Dendrogram-Slider X-Axis. Use to adjust dendrogram cluster sizes - slide left for larger clusters and right for smaller clusters. Hover mouse over grey trapezoid to get information on each cluster.
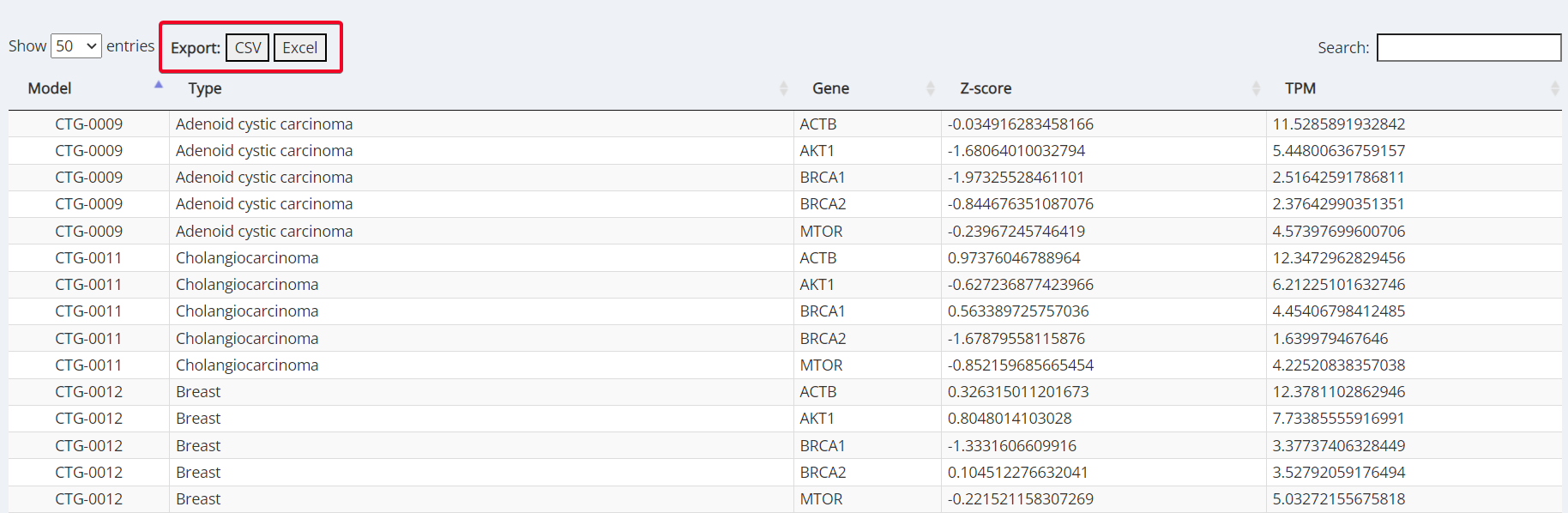
Clustergrammer Table
View all the models within the heatmap as a table for each gene of interest, z-score and TPM value. This table can be exported as a CSV or an Excel File by clicking the button at the top left of the table (highlighted in red).
Available Data
Champions PDX RNASeq
Champions PDX Proteomics
Champions PDX Phosphoproteomics
Champions PDX Kinase
Champions Cell Lines
TCGA
GEO
CPTAC
Your Data
Key Terms
Clustering
Clustering is the process of grouping the data into clusters or classes, so that objects within a cluster have high similarity in comparison to one another but are dissimilar to objects in other clusters.
TPM
Transcripts Per Million (TPM) is a normalization method for RNA-seq. You can interpret this as "for every 1,000,000 RNA molecules in the RNA-Seq sample, x came from this gene/transcript."
Z-Score
Z-Scores are calculated by subtracting the overall average gene abundance from the raw expression for each gene, and dividing that result by the standard deviation (SD) of all of the measured counts across all samples or across the selected criteria depending on which you choose.
TGI
Tumor Growth Inhibition (TGI) rate is a common metric used to quantify agent response of treatments compared to the control. It is calculated using the formula (1 – (mean volume of treated tumors)/(mean volume of control tumors)) × 100%.
Common Analysis with the Clustergrammer
Visualizing RNA-Seq for a Pathway of Interest
- Click the pathways icon after 'Genes of interest'
- Find your pathway of interest and click 'Select genes'
- Select any gene mutations of interest. These do not need to match the RNA-Seq genes.
- Select your desired dataset.
- Select a specific cancer type, if desired. A specific cancer type is required for all datasets other than Champions PDX.
- Click 'View'
Exploring Champions Proteomics/Kinase Data
- Input your genes or proteins of interest in the 'Genes of interest' text bar.
- Select either 'Champions PDX Proteomics', 'Champions PDX Phosphoproteomics' or 'Champions PDX Kinase' dataset.
- Select a specific cancer type. A specific cancer type is required for all datasets other than Champions PDX.
- Click 'View'
View Available In-Vivo SOC Response Data
- Under 'Clustering Type' select 'Agent Response.
- Select 'Champions PDX RNASeq' dataset.
- Select a specific cancer type, if desired. If not keep as all cancer types.
- Select a specific test agent, if desired. If not keep as all available.
- Click 'View'
Explore Public Data RNAseq
- Input your genes or proteins of interest in the 'Genes of interest' text bar.
- Select either 'TCGA', 'GEO' or 'CPTAC' dataset.
- Select a specific cancer type. A specific cancer type is required for all datasets other than Champions PDX.
- Click 'View'
Compare Pre & Post-Study Sequencing
Coming Soon!!
References and Acknowledgements
Clustergrammer is based on the work of Fernandez et al: Fernandez, N. F. et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data 4:170151 doi: 10.1038/sdata.2017.151 (2017).
Updated almost 2 years ago
