Oncoprint
Explore whole exome sequencing data.
General Description
Our Oncoprint tool aims to help users visualize genetic alterations in user defined genes across Champions PDX models.
Oncoprint Input
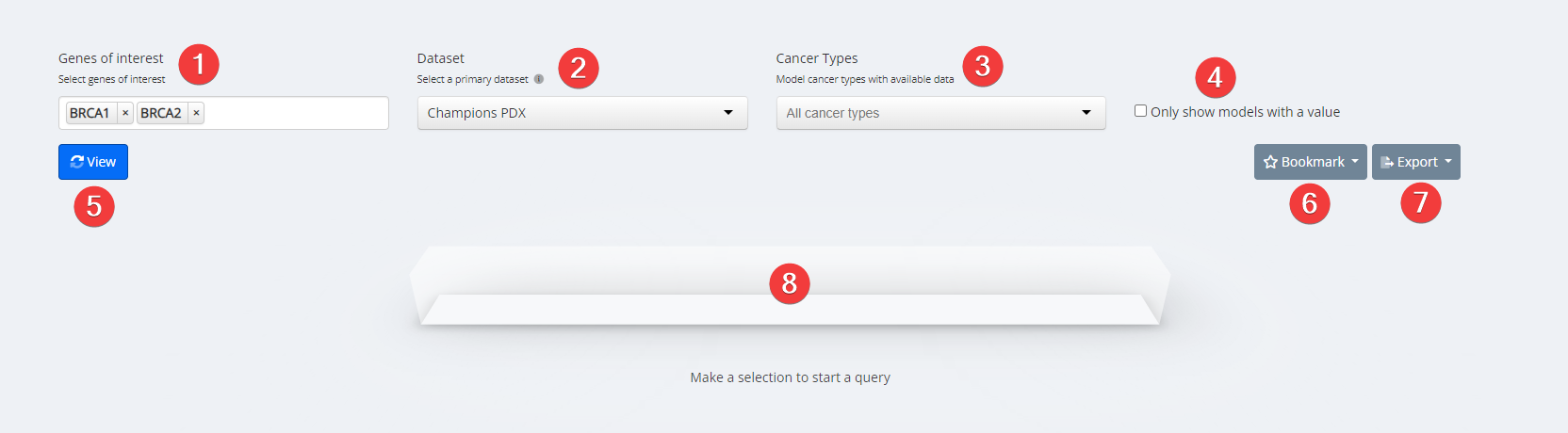
Oncoprint Input
- Gene of Interest. Select your gene of interest from the dropdown. Please Note: You must choose at least one gene in order to run visualization.
- Dataset. Choose from the dropdown menu between Champions PDX & Cell Lines, Online Database such as TCGA, GEO, CPTAC, or your own data previously uploaded to Lumin.
- Cancer Types. Select cancer type of interest from the dropdown menu or leave blank for all cancer types.
- Checkbox. Click the checkbox to only show models with a value.
- View.
- Bookmark. Save an analysis or load from an existing analysis. Please Note: As we are continually updating our data the visualization may change slightly from the time it was first analyzed. Users can also manage all bookmarks under "My Bookmarks" in Account Settings at the top right of any Lumin page.
- Export. Analysis can be exported as a CSV file, PNG image or into Lumin Workspaces.
- Loading Task Bar. Building your requested visualization may take some time. The status of your request will be displayed here.
Oncoprint Visualization
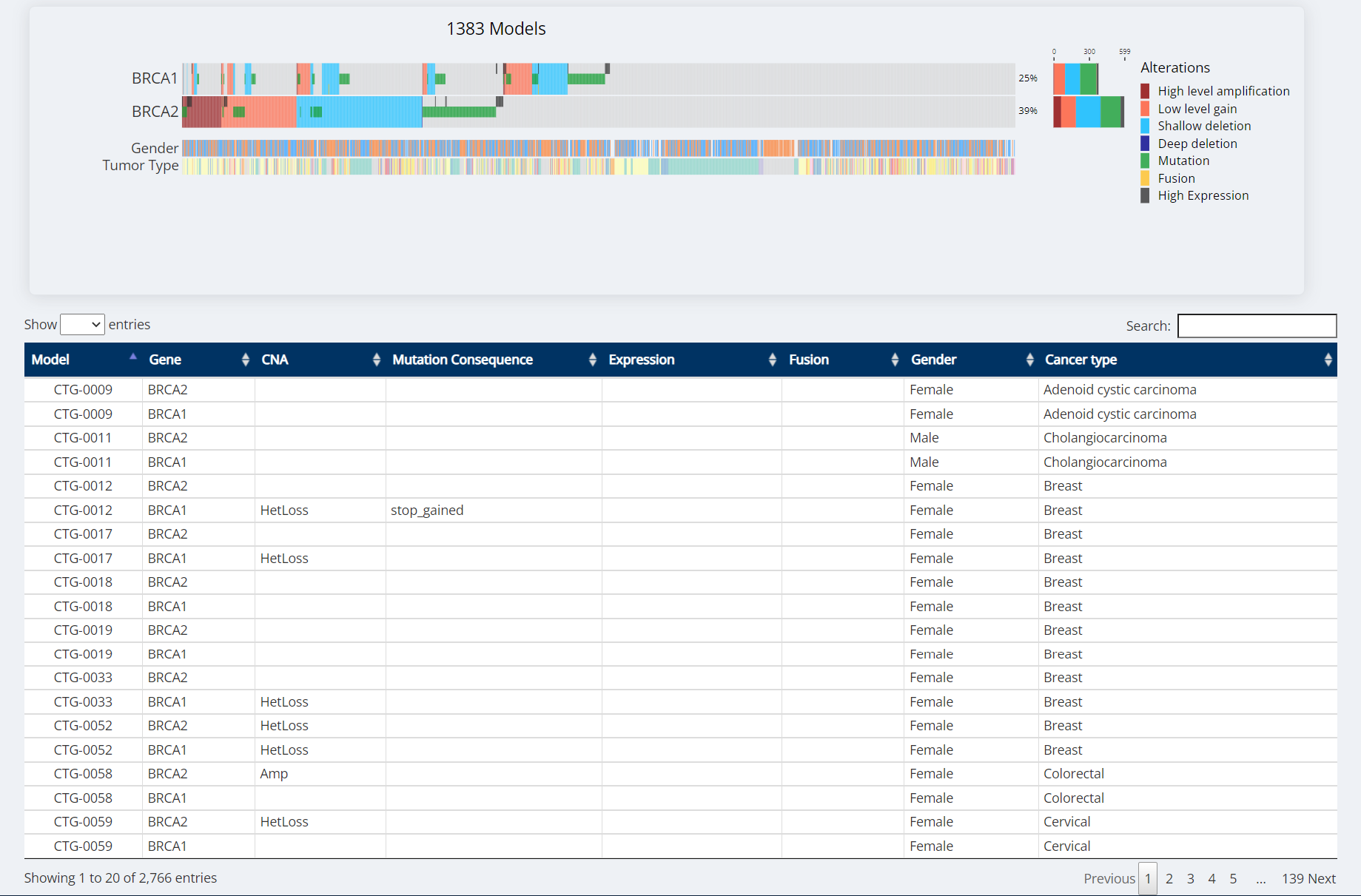
Oncoprint Visualization
CNA Thresholds
| Alteration | Copy Number |
|---|---|
| Deep Deletion (HomDel) | 0 |
| Shallow Deletion (HetLoss) | 1 |
| Low Level Gain (Gain) | 3 |
| High Level Amplification (Amp) | > =4 |
Available Data
Champions PDX
Champions Cell Lines
TCGA
GEO
CPTAC
Your Data
Key Terms
CNA - Copy Number Alteration
Common Analysis with Oncoprint
Coming Soon!
References and Acknowledgements
Updated over 2 years ago
