Dependency Heatmap
General Description
Dependency Heatmap Input
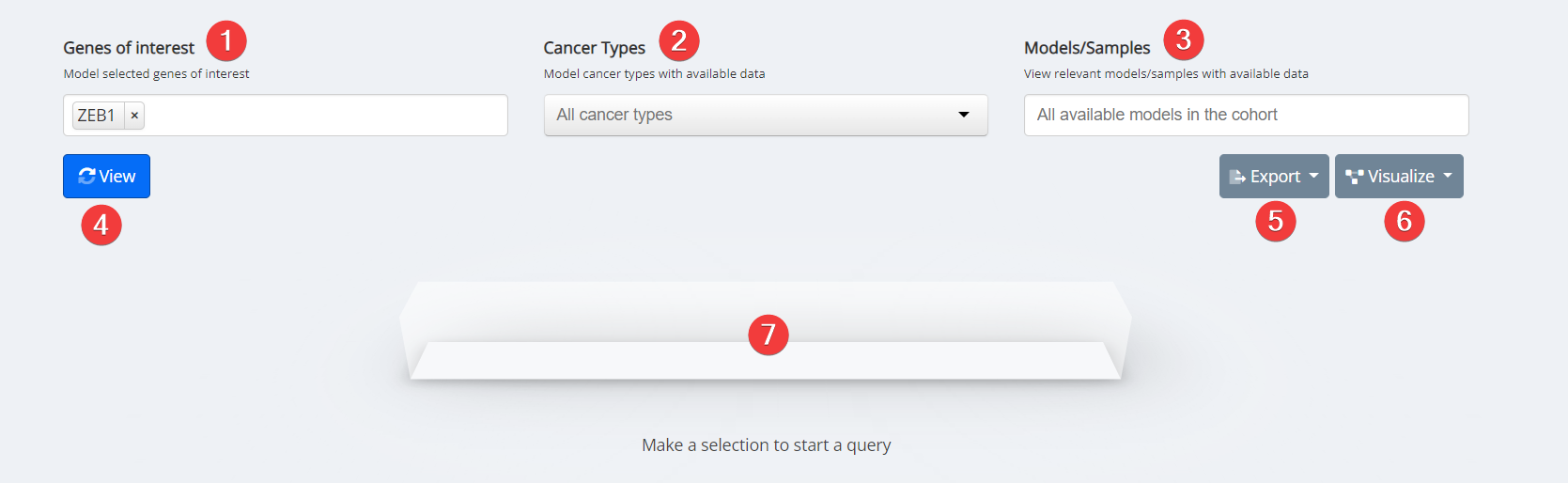
Dependency Heatmap Input
- Genes of Interest. Select your genes of interest. Only options with available data for your other selections will be clickable. Options that are greyed out have no available data for your other selections.
- Cancer Type. Select cancer type of interest from the dropdown menu or leave blank for all cancer types.
- Model/Sample. Select specific models with available data or leave blank for all available models with data.
- View.
- Export. Analysis can be exported as a PNG image.
- Visualize. Data inputs can be populated and visualized in Oncoprint, Mutation Mapper, and High/Low Expression.
- Loading Task Bar. Building your requested visualization may take some time. The status of your request will be displayed here.
Dependency Heatmap Visualization
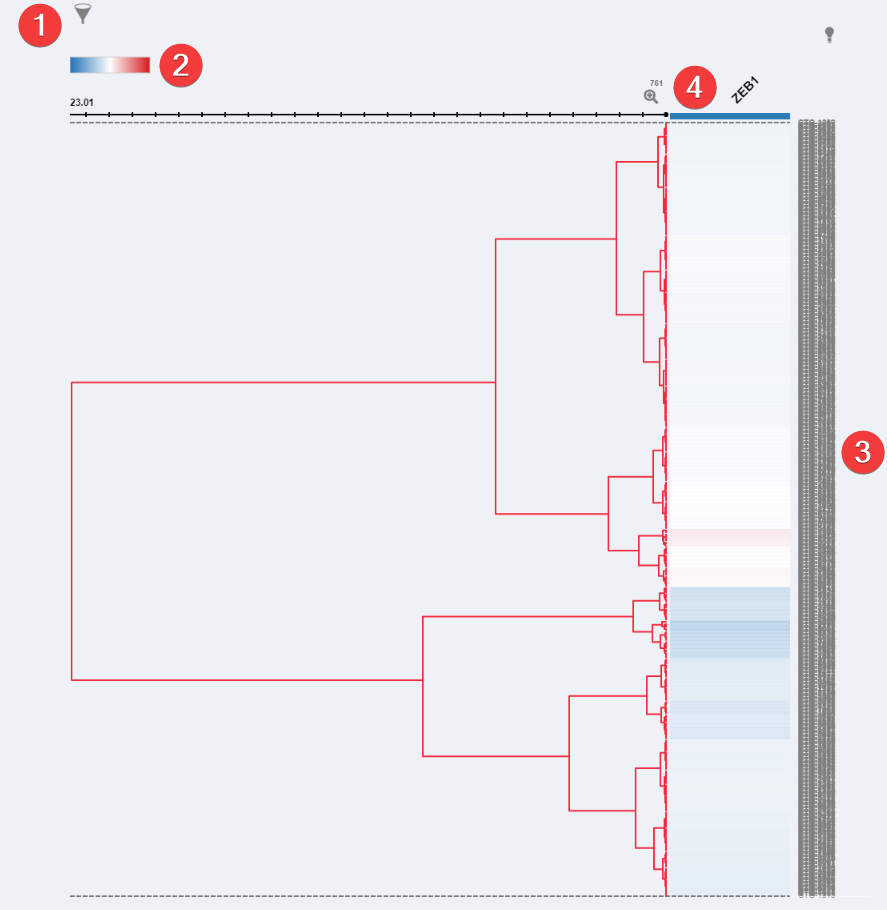
Dependency Heatmap Visualization
- Filter. When genes are selected you can filter which are visible.
- Color Settings. Change the heatmap data colors.
- Dendrogram. Shows each model with data for a chosen genes and the z-score for each of the models.
- Zoom. After selecting a row you can click the 'Zoom' Icon to have a closer look at the models within that row.
A machine learning model (gradient boost machine) was trained using the Depmap-CCLE data to predict the survival dependencies of cells on a list of 200 genes. The prediction works based on the transcriptional and mutational profile of cells, and outputs the predicted dependencies for each of 200 genes. As per the original Depmap-CCLE data, the values range from negative values to positive values, with strong negative values predicting reduced viability, and
strong positive values predicting increased viability, with the ablation of the gene.
Key Terms
Common Analysis
Coming Soon!!
References and Acknowledgements
Depmap.org. 2022. DepMap: The Cancer Dependency Map Project at Broad Institute. [online] Available at: https://depmap.org/portal/ccle/ [Accessed 2022].
Updated over 2 years ago
