Molecular Signatures
Explore molecular signatures for Champions' PDX models, public datasets, or your own custom models.
General Description
Our signatures tool aims to allow users to upload datasets to build their own gene signatures or access several precomputed molecular signatures that leverage data from Champions PDX models, TCGA, Achilles and CPTAC databases.
Correlations for each molecular signature are calculated using by Pearson correlation.
Molecular Signatures Input
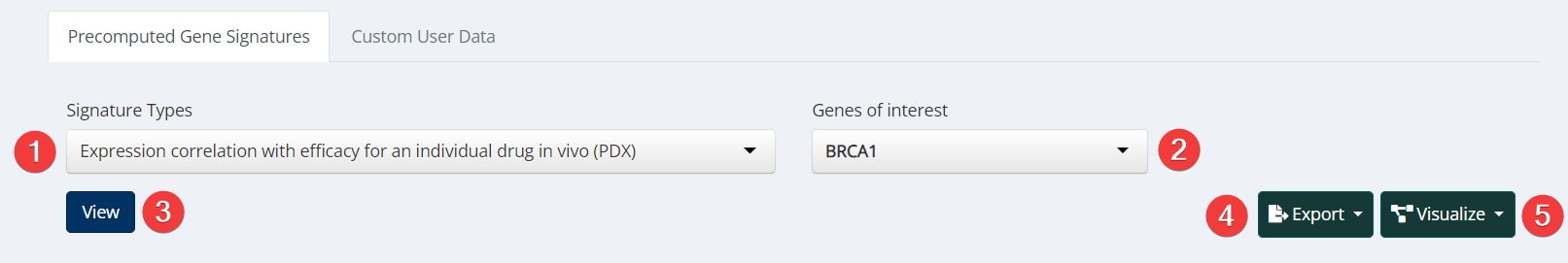
Molecular Signatures Input
- Signature Type Input. Select your data of interest from the following options in the dropdown menu - Molecular Signatures (PDX), Correlation with Survival (TCGA), Genetic Dependency (Achilles) and Proteomics (CPTAC).
- Gene Input. Select your genes or proteins of interest. Only options with available data for your other selections will be clickable. Options that are greyed out have no available data for your other selections.
- View.
- Export. Analysis can be exported as a CSV file, PNG image or into Lumin Workspaces.
- Visualize. Data inputs can be populated and visualized in Clustering Heatmap, Target and Biomarker Discovery, Oncoprint, Mutation mapper, and High/Low expression.
Molecular Signatures Visualization
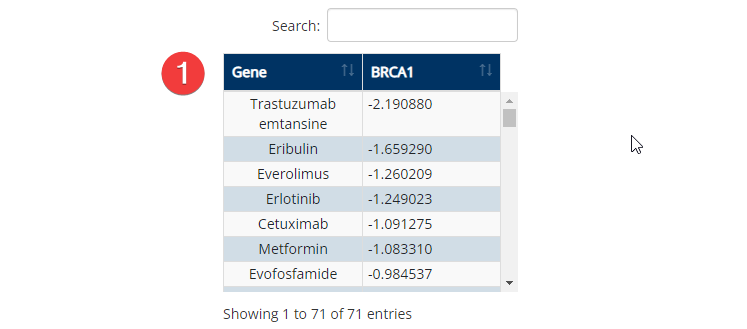
Molecular Signatures Visualization
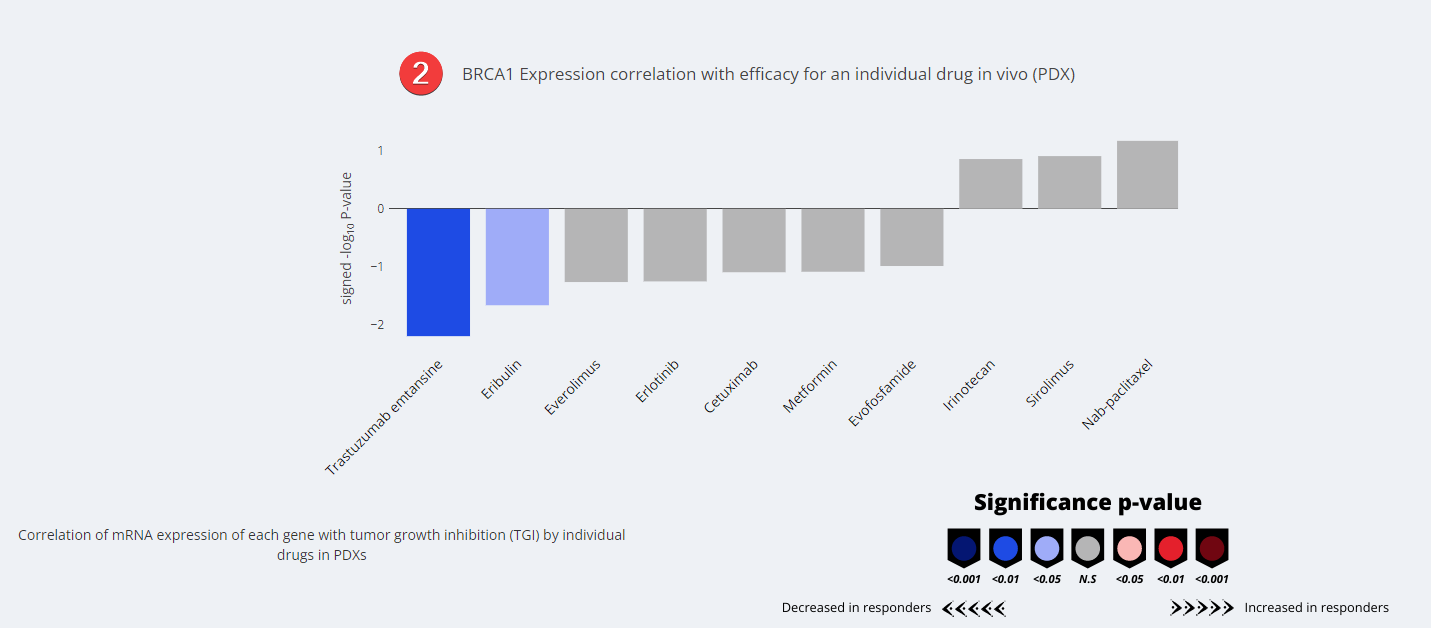
Molecular Signatures Visualization
- Full List. Lists the agents with efficacy data available for the gene selected in the 'Genes of Interest' input.
- Waterfall Plot. Showing a selected few of the agents plotted based on their signed -log10 p-value.
Available Data
Champions PDX
TCGA
Achilles
CPTAC
Your Data
Key Terms
Expression Change After Treatment With:
The Champions PDX bank was derived from human patient tumors prior to (treatment-naïve) or after undergoing standard-of-care therapy (post-therapy). Molecular data (whole-exome and RNA sequencing) from these PDXs was obtained after passaging (2-3 passages) in mice. This gene signature was calculated based on the differential expression of the user specified gene in PDXs obtained from post-therapy vs. treatment-naïve patient tumors. Thus, this signature reflects the gene expression landscape of the indicated tumors after undergoing standard-of-care therapy with the indicated drugs.
The individual values are sign-corrected log10p-values of gene expression differences. E.g. a value of -2, (representing a p-value of 0.01), for gene “A” indicates that the expression of gene “A” is reduced in patient cancer samples after receiving the standard-of-care therapies described relative to treatment-naïve samples. Conversely, a value of 2, (representing a p-value of 0.01),indicates increased expression of gene “A” in patient cancer samples after receiving the standard-of-care therapies described relative to treatment-naïve samples.
Expression correlation with clinical response:
The Champions PDX bank was derived from human patient tumors prior to (treatment-naïve) or after undergoing standard-of-care therapy (post-therapy). Molecular data (whole-exome and RNA sequencing) from these PDXs was obtained after passaging (2-3 passages) in mice. Many patients underwent further therapy after sample collection for PDX, and their clinical response status to these therapies were obtained by Champions. This signature is calculated based on the differential expression of the user defined gene in the PDXs whose original donor patients went on to respond to the indicated therapy vs. PDXs whose original donor patients did not respond to the indicated therapy. Thus, this signature reflects the pre-treatment gene expression landscape in the therapy-responsive vs non-responsive tumors.
The individual values are sign-corrected log10p-values of expression difference. E.g. a value of -2, (representing a p-value of 0.01),for gene “A” indicates that the expression of gene “A” is reduced in PDXs whose donor patients later responded to therapy relative to patients that did not respond that same therapy. Conversely, a value of 2, (representing a p-value of 0.01),indicates increased expression in PDXs whose donor patients later responded to therapy relative to patients that did not respond that same therapy.
Expression correlation with overall survival (TCGA):
The Cancer Genome Atlas (TCGA) data contain molecular data from >11,000 patient tumors, along with their overall survival data post-diagnosis. This signature reflects correlation of a gene’s mRNA expression with the overall survival in each of the TCGA cancer indications described.
The individual values are sign-corrected log10p-values of correlation. For example, a value of -2, (representing a p-value of 0.01),for gene “A” indicates that when the expression of gene “A” is high patient outcome is better with longer survival, whereas a positive 2value, (representing a p-value of 0.01),indicates the opposite: high expression of gene “A” predicts worse out come or poorer survival.
Mutation enrichment after treatment with:
The Champions PDX bank was derived from human patient tumors prior to (treatment-naïve) or after undergoing standard-of-care therapy (post-therapy). Molecular data (whole-exome and RNA sequencing) from these PDXs was obtained after passaging (2-3 passages) in mice. This gene signature was calculated based on the differential mutation of the user specified gene in PDXs obtained from post-therapy vs. treatment-naïve patient tumors. Thus, this signature reflects the post-therapy mutational landscape of the indicated tumors after undergoing standard-of-care therapy with the indicated drugs.
The individual values are sign-corrected log10p-values of differential mutation enrichment. E.g. a value of -2, (representing a p-value of 0.01),for gene “A” indicates that gene “A” is less frequently mutated post-therapy relative to treatment-naïve PDXs (i.e. mutations in “A” are depleted after therapy). Conversely, a value of 2, (representing a p-value of 0.01),indicates that the gene “A” is more frequently mutated post-therapy relative to treatment-naïve PDXs (i.e. mutations in “A” are gained after therapy).
Mutation correlation with overall survival(TCGA):
The Cancer Genome Atlas (TCGA) data contain molecular data from >11,000 patient tumors, along with their overall survival data post-diagnosis. This signature reflects correlation of a gene’s mutation with overall survival for each of the TCGA cancer indications described.
The individual values are sign-corrected log10p-values of correlation. E.g. a value of -2, (representing a p-value of 0.01),for gene “A” indicates that when the gene “A” is mutated the patient outcome tends to be better(longer survival), whereas a positive 2, (representing a p-value of 0.01),value indicates the opposite: mutation of gene “A” predicts worse patient outcome (shorter survival).
Mutation correlation with clinical response:
The Champions PDX bank was derived from human patient tumors prior to (treatment-naïve) or after undergoing standard-of-care therapy (post-therapy). Molecular data (whole-exome and RNA sequencing) from these PDXs was obtained after passaging (2-3 passages) in mice. Many patients underwent further therapy, and their clinical response status to these therapies were obtained by Champions. This signature is calculated based on the differential mutation of the user defined gene in the PDXs whose original donor patients went on to respond to the indicated therapy vs. PDXs whose original donor patients did not respond to the indicated therapy. Thus, this signature reflects the pre-treatment genetic mutational landscape in the therapy-responsive vs non-responsive tumors.
The individual values are sign-corrected log10p-values of mutation correlation. E.g. a value of -2, (representing a p-value of 0.01),for gene “A” indicates that mutation of gene “A” is less frequent, or depleted) in PDXs whose donor later responded to the therapy relative to patients that did not respond to the same therapy. Conversely, a value of 2,(representing a p-value of 0.01),indicates increased mutation rates for gene “A” in PDXs whose donor later responded to the therapy relative to patients that did not respond to the same therapy.
Expression correlation with efficacy for (PDX):
The Champions PDX bank is comprised of a collection of 1000+ low-passage PDXs derived from human tumor samples. As part of our internal research and development, we performed over 3,400 in vivo PDX efficacy studies and obtained response rates of our PDX models to a number standard of care therapies. This signature was calculated by correlating the specified gene’s expression across PDX models with efficacy (tumor growth inhibition index) to the standard of care rugs described. Thus, this signature reflects the gene expression profile that predicts in vivo efficacy to standard of care drugs tested in our PDX models.
The individual values are sign-corrected log10p-values of expression difference. E.g. a value of -2, (representing a p-value of 0.01),for gene “A” indicates that high expression of gene “A” predicts poor efficacy (resistance) to the specified standard of care agent in PDX models. Conversely, a value of2, (representing a p-value of 0.01),indicates the opposite: high expression of gene “A” predicts better efficacy to standard of care drugs listedin PDX models. For instance, the gene EGFR has a strongly positive value of 9.6 (corresponding to P < 10-9) for Cetuximab, which is a mAb against EGFR, as expected, indicating that PDXs with high expression of EGFR are particularly sensitive to Cetuximab.
Correlation of genetic dependency with (Achilles):
The project “Achilles” led by Broad Institute aims to delineate the map of genetic dependencies in cancer cell lines. The current dataset comprises genetic dependency values for 18,000+ genes across 739 cancer cell lines. This gene signature was calculated by correlating the expression, mutation or copy number variations of genes across cell lines with the genetic dependency of user-defined gene(s)across the same cell lines.
*The individual values shown are statistically significant Pearson’s r-values of correlation, which range from -1 for a perfect negative correlation to 1 for a perfect positive correlation. A negative correlation for gene “A”(the gene input by the user)with the expression of gene “B” indicates that high expression of “B” predicts less viability (higher toxicity) of cells upon the knock-out of gene “A”. Conversely, a positive value for gene “A” (the gene input by the user)with the expression of gene “B” indicates that high expression of “B” predicts better viability of cells upon knock-out of “A”.
Common Analysis with Molecular Signatures
Coming Soon!
References and Acknowledgements
Waterfall plots produced by Plotly. The front end for ML and data science models. Plotly. (n.d.). Retrieved 2022, from https://plotly.com/
Updated over 2 years ago
