Pathways Heatmap
Gene Set Enrichment Analysis
General Description
Our Gene Set Enrichment Pathways tool aims to provide a standard ssGSEA enrichment score for each of our PDX models.
Pathways Heatmap Input
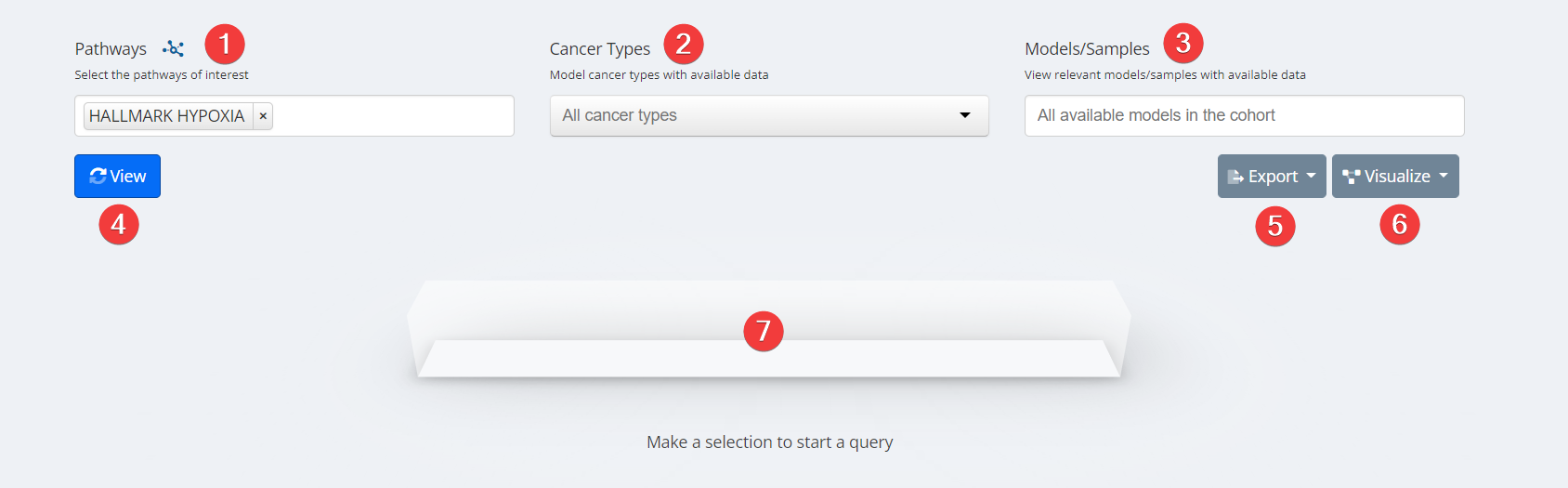
Pathways Heatmap Input
- Pathways. Select your pathway of interest.
- Cancer Type. Select cancer type of interest from the dropdown menu or leave blank for all cancer types.
- Model/Sample. Select specific models with available data or leave blank for all available models with data.
- View.
- Export. Analysis can be exported as a PNG image.
- Visualize. Data inputs can be populated and visualized in Oncoprint, Mutation Mapper, and High/Low Expression.
- Loading Task Bar. Building your requested visualization may take some time. The status of your request will be displayed here.
Pathways Heatmap Visualization
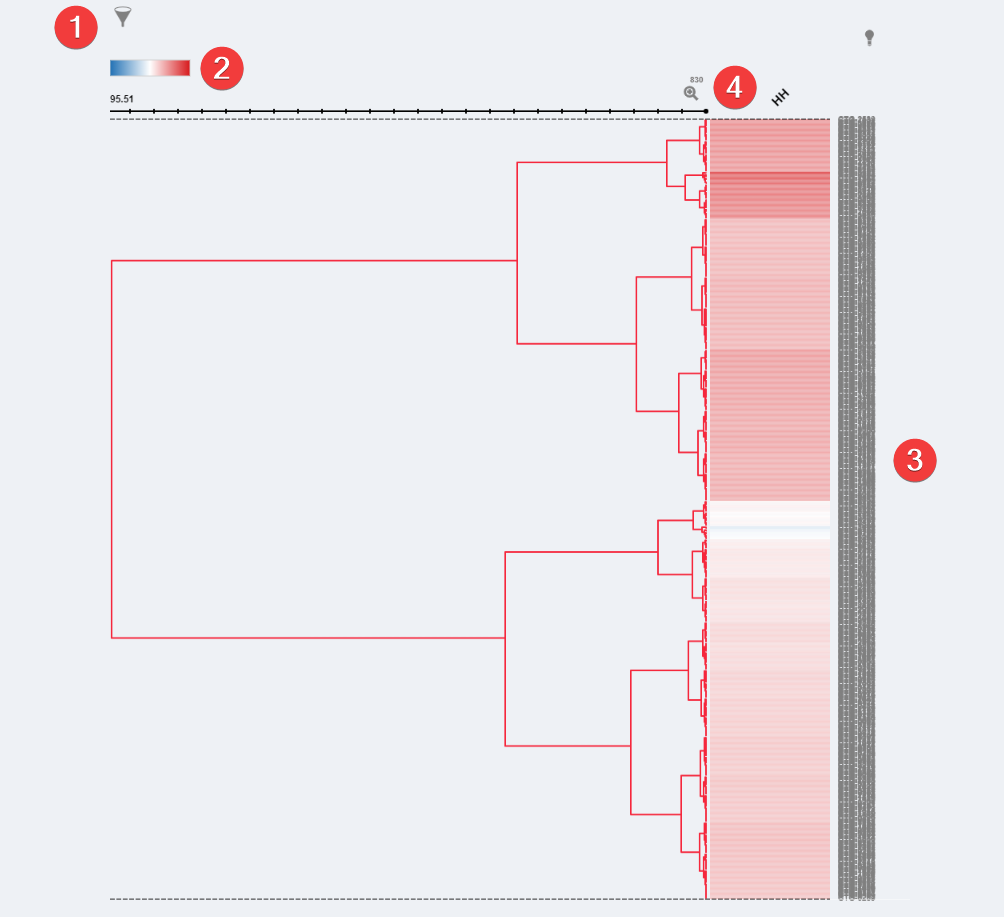
Pathways Heatmap Visualization
- Filter. When multiple pathways are selected you can filter which are visible.
- Color Settings. Change the heatmap data colors.
- Dendrogram. Shows each model with data for a chosen pathway and the z-score for each of the models.
- Zoom. After selecting a row you can click the 'Zoom' Icon to have a closer look at the models within that row.
The Z-score values shown provide a standardized metric of a specified gene set enrichment score across all of the samples in our database. A higher Z-score value shown for a specified model would represent higher than "average" expression of the gene set within that model when compared to all models in our database. For example, a sample with a Z-score of 2 indicates this sample has a score 2 standard deviation higher than the average score in the database.
Key Terms
ssGSEA - Single Sample Gene Set Enrichment Analysis
Common Analysis with Pathways Heatmap
Coming Soon!!
References and Acknowledgements
Updated over 2 years ago
